Paper of the Month - January 2026
selected by the BMAS Scientific Board
Multifaceted bone response to immunomodulatory magnesium implants: Osteopromotion at the interface and adipogenesis in the bone marrow
- 1Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy, University of Gothenburg, Sweden.
- 2Biomaterials Group, Materials Design Division, Faculty of Materials Science and Engineering, Warsaw University of Technology, Poland.
- 3Institute of Metallic Biomaterials, Helmholtz-Zentrum Hereon, Geesthacht, Germany.
- 4Department of Biomedical Dental Sciences, College of Dentistry, Imam Abdulrahman Bin Faisal University, Dammam, Saudi Arabia.
- 5Department of Biomaterials, Institute of Clinical Sciences, Sahlgrenska Academy, University of Gothenburg, Sweden.
Correspondence:Peter Thomsen: peter.thomsen@biomaterials.gu.se
Biomaterials. 2025;314:122779.
PMID: 39305536 | DOI: 10.1016/j.biomaterials.2024.122779
Key Findings
Orthopedic implants for fracture repair interact closely with the bone marrow and BMAT, and the initial phase of bone healing is key to the successful integration of biodegradable Mg implants. The study by Ben Amara et al., published in Biomaterials (2025), revealed that both high-purity Mg implants and clinical-grade, rare-earth-alloyed implants (MgYREZr) elicit proangiogenic gene regulation coupled with osteoclastogenesis activation and persistent low-grade inflammation. Moreover, Mg degradation triggered adipogenic pathways in the bone marrow near the implant, with bone marrow adipocytes identified via semiautomated morphometry in the peri-implant region adjacent to CD68-expressing cells. This study suggests a degradation-dependent dichotomous organization of peri-implant tissues, underscoring the need to focus not only on the osseointegration of Mg implants in patients but also on the fate of surrounding bone marrow.
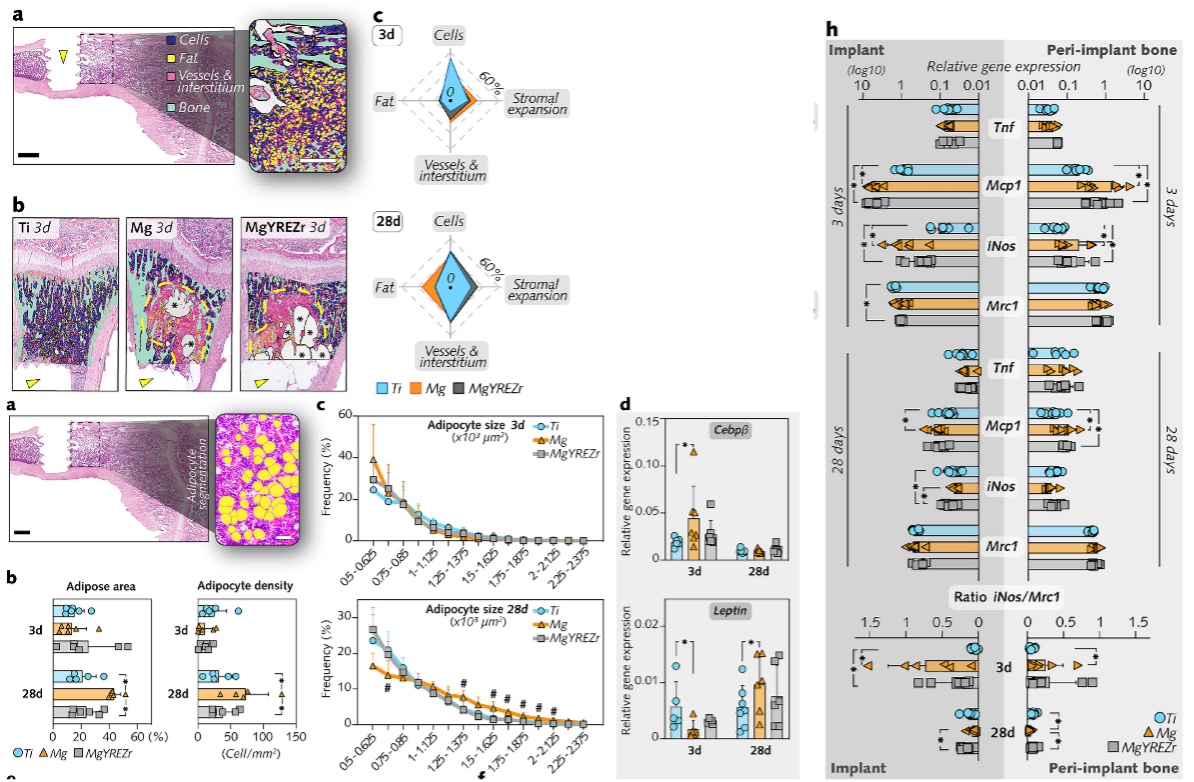
Figure: Mg degradation increased the number of bone marrow adipocytes in the physeal region distant to the interface without altering the trabecular bone architecture.
